Bioinformatics: new tools and applications in life science and personalized medicine

While we have a basic understanding of the functioning of the gene when coding sequences of specific proteins, we feel the lack of information on the role that DNA has on specific diseases or functions of thousands of proteins that are produced. Bioinformatics combines the methods used in the collection, storage, identification, analysis, and correlation of this huge and complex information. All this work produces an “ocean” of information that can only be “sailed” with the help of computerized methods. The goal is to provide scientists with the right means to explain normal biological processes, dysfunctions of these processes which give rise to disease and approaches that allow the discovery of new medical cures. Recently, sequencing platforms, a large scale of genomes and transcriptomes, have created new challenges not only to the genomics but especially for bioinformatics. The intent of this article is to compile a list of tools and information resources used by scientists to treat information from the massive sequencing of recent platforms to new generations and the applications of this information in different areas of life sciences including medicine.
Key points
• Biological data mining
• Omic approaches
• From genotype to phenotype
This is a preview of subscription content, log in via an institution to check access.
Access this article
Subscribe and save
Springer+ Basic
€32.70 /Month
- Get 10 units per month
- Download Article/Chapter or eBook
- 1 Unit = 1 Article or 1 Chapter
- Cancel anytime
Buy Now
Price includes VAT (France)
Instant access to the full article PDF.
Rent this article via DeepDyve
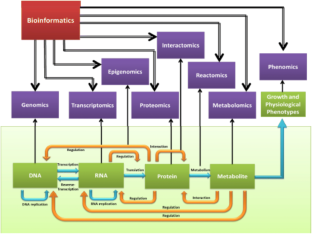
Similar content being viewed by others

Bioinformatics
Chapter © 2021

Bioinformatics
Chapter © 2014

Bioinformatics advances biology and medicine by turning big data troves into knowledge
Article 22 March 2017
References
- Allen JE, Salzberg SL (2005) JIGSAW: integration of multiple sources of evidence for gene prediction. Bioinformatics 21:3596–3603 ArticleCASPubMedGoogle Scholar
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410 ArticleCASPubMedGoogle Scholar
- Bader GD, Betel D, Hogue CW (2003) BIND: the biomolecular interaction network database. Nucleic Acids Res 31(1):248–250. https://doi.org/10.1093/nar/gkg056ArticleCASPubMedPubMed CentralGoogle Scholar
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S (2004a) Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 340:783–795 ArticlePubMedGoogle Scholar
- Bendtsen JD, Jensen LJ, Blom N, Von Heijne G, Brunak S (2004b) Feature-based prediction of non-classical and leaderless protein secretion. Protein Eng Des Sel 17(4):349–356 ArticleCASPubMedGoogle Scholar
- Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW (2010) GenBank. Nucleic Acids Res 38(Database issue):D46–D51. https://doi.org/10.1093/nar/gkp1024ArticleCASPubMedGoogle Scholar
- Bonfield JK, Whitwham A (2010) Gap5--editing the billion fragment sequence assembly. Bioinformatics 26(14):1699–1703 ArticleCASPubMedPubMed CentralGoogle Scholar
- Burge C, Karlin S (1997) Prediction of complete gene structures in human genomic DNA. J Mol Biol 268:78–94 ArticleCASPubMedGoogle Scholar
- Buza T, Tonui T, Stomeo F, Tiambo C, Katani R, Schilling M, Lyimo B, Gwakisa P, Cattadori IM, Buza J, Kapur V (2019) iMAP: an integrated bioinformatics and visualization pipeline for microbiome data analysis. BMC Bioinformatics 20:374 ArticlePubMedPubMed CentralGoogle Scholar
- Bystroff C, Shao Y (2002) Fully automated ab initio protein structure prediction using I-SITES, HMMSTR and ROSETTA. Bioinformatics 18(Suppl 1):S54–S61 ArticlePubMedGoogle Scholar
- Chen C, Natale DA, Finn RD, Huang H, Zhang J, Wu CH, Mazumder R (2011) Representative proteomes: a stable, scalable and unbiased proteome set for sequence analysis and functional annotation. PLoS One 6(4):e18910 ArticleCASPubMedPubMed CentralGoogle Scholar
- Chen M, Hofestädt R, Taubert J (2019) Integrative bioinformatics: history and future. Journal of Integrative Bioinformatics 16. https://doi.org/10.1515/jib-2019-2001
- Chordia N, Kumar A (2018) Bioinformatics in drug discovery. SF Protein Sci J 1:1 Google Scholar
- Chou K, Shen H (2010) A new method for predicting the subcellular localization of eukaryotic proteins with both single and multiple sites: Euk-mPLoc 2.0. PLoS One
- Dawson NL, Lewis TE, Das S, Lees JG, Lee D, Ashford P, Orengo CA, Sillitoe I (2017) CATH: an expanded resource to predict protein function through structure and sequence. Nucleic Acids Res 45:D289–D295. https://doi.org/10.1093/nar/gkw1098ArticleCASPubMedGoogle Scholar
- Di-Lena P, Wu G, Martelli PL, Casadio R, Nardini C (2013) MIMO: an efficient tool for molecular interaction maps overlap. BMC Bioinformatics 14:159 10.1186/1471-2105-14-159. 10.1093/bioinformatics/btn596 ArticlePubMedPubMed CentralGoogle Scholar
- Drozdetskiy A, Cole C, Procter J, Barton GJ (2015) JPred4: a protein secondary structure prediction server. Nucleic Acids Res 43(Web Server issue):W389–W394. https://doi.org/10.1093/nar/gkv332ArticleCASPubMedPubMed CentralGoogle Scholar
- El-Gebali S, Mistry J, Bateman A, Eddy SR, Luciani A, Potter SC, Qureshi M, Richardson LJ, Salazar GA, Smart A, Sonnhammer ELL, Hirsh L, Paladin L, Piovesan D, Tosatto SCE, Finn RD (2019) The Pfam protein families database in 2019: nucleic acids res. https://doi.org/10.1093/nar/gky995
- Finn RD, Clements J, Eddy SR (2011) HMMER web server: interactive sequence similarity searching. Nucleic Acids Res 39:W29–W37 ArticleCASPubMedPubMed CentralGoogle Scholar
- Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I (2002) VISTA: computational tools for comparative genomics. Nucleic Acids Res 32(Web Server issue):W273–W279 Google Scholar
- Ganesan N, Bennett NF, Velauthapillai M, Pattabiraman N, Squier R, Kalyanasundaram B (2005) Web-based interface facilitating sequence-to-structure analysis of BLAST alignment reports. Biotechniques 39(186):188 Google Scholar
- Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A (2005) In: The proteomics protocols handbook. Protein identification and analysis tools on the ExPASy server. Springer, pp 571-607
- Goldberg T, Hamp T, Rost B (2012) LocTree2 predicts localization for all domains of life. Bioinformatics 28(18):i458–i465. https://doi.org/10.1093/bioinformatics/bts390ArticleCASPubMedPubMed CentralGoogle Scholar
- Goldman D, Domschke K (2014) Making sense of deep sequencing. Int J Neuropsychopharmacol 17(10):1717–1725. https://doi.org/10.1017/S1461145714000789ArticleCASPubMedGoogle Scholar
- Günther S, Kuhn M, Dunkel M, Campillos M, Senger C, Petsalaki E, Ahmed J, Urdiales EG, Gewiess A, Jensen LJ, Schneider R, Skoblo R, Russell RB, Bourne PE, Bork P, Preissner R (2008) SuperTarget and Matador: resources for exploring drug-target relationships. Nucleic Acids Res 36(Database issue):D919–D922. https://doi.org/10.1093/nar/gkm862ArticleCASPubMedGoogle Scholar
- Heather JM, Chain B (2016) The sequence of sequencers: the history of sequencing DNA. Genomics 107(1):1–8. https://doi.org/10.1016/j.ygeno.2015.11.003ArticleCASPubMedGoogle Scholar
- Horler RS, Butcher A, Papangelopoulos N, Ashton PD, Thomas GH (2009) EchoLOCATION: an in silico analysis of the subcellular locations of Escherichia coli proteins and comparison with experimentally derived locations. Bioinformatics 25(2):163–166 ArticleCASPubMedGoogle Scholar
- Hunt SE, McLaren W, Gil L, Thormann A, Schuilenburg H, Sheppard D, Parton A, Armean IM, Trevanion SJ, Flicek P, Cunningham F (2018) Ensembl variation resources. Database Volume 2018 https://doi.org/10.1093/database/bay119
- Ideker T, Kelley, Shamir R, Karp R (2004) Identification of protein complexes by comparative analysis of yeast and bacterial protein interaction data Proceedings: RECOMB 2004, pp. 282-289; J Comput Biol 12: 835–846, 2005
- Jiang P, Sun K, Lun FMF, Guo AM, Wang H, Chan KCA, Rossa WK, ChiuY M, Lo D, Sun H (2014) Methy-Pipe: an integrated bioinformatics pipeline for whole genome bisulfite sequencing data analysis. PLoS One 9(6):e100360. https://doi.org/10.1371/journal.pone.0100360ArticlePubMedPubMed CentralGoogle Scholar
- Kalendar R, Khassenov B, Ramankulov Y, Samuilova O, Ivanov KI (2017a) FastPCR: an in silico tool for fast primer and probe design and advanced sequence analysis. Genomics 109:312–319 ArticleCASPubMedGoogle Scholar
- Kalendar R, Muterko A, Shamekova M, Zhambakin K (2017b) In silico PCR tools a fast primer, probe and advanced searching. Methods Mol Biol 1620:1–31. https://doi.org/10.1007/978-1-4939-7060-5_1ArticleCASPubMedGoogle Scholar
- Kalendar R, Tselykh T, Khassenov B, Ramanculov EM (2017c) Introduction on using the FastPCR software and the related Java web tools for PCR, in silico PCR, and oligonucleotide assembly and analysis. Methods Mol Biol 1620:33–64. https://doi.org/10.1007/978-1-4939-7060-5_2ArticleCASPubMedGoogle Scholar
- Källberg M, Wang H, Wang S, Peng J, Wang Z, Lu H, Xu J (2012) Template-based protein structure modeling using the RaptorX web server. Nat Protoc 7:1511–152253 ArticlePubMedPubMed CentralGoogle Scholar
- Kalvari I, Nawrocki EP, Argasinska J, Quinones-Olvera N, Finn RD, Bateman A, Petrov AI (2018) Non-coding RNA analysis using the Rfam database. Curr Protoc Bioinformatics 62(1):e51. https://doi.org/10.1002/cpbi.51ArticleCASPubMedPubMed CentralGoogle Scholar
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10:845–858 ArticleCASPubMedPubMed CentralGoogle Scholar
- Khan FA, Phillips CD, Baker RJ (2014) Timeframes of speciation, reticulation, and hybridization in the bulldog bat explained through phylogenetic analyses of all genetic transmission elements. Syst Biol 63:96–110 ArticleCASPubMedGoogle Scholar
- Kinjo AR, Suzuki H, Yamashita R, Ikegawa Y, Kudou T, Igarashi R, Kengaku Y, Cho H, Standley MD, Nakagawa A, Nakamura H (2012) Protein Data Bank Japan (PDBj): maintaining a structural data archive and resource description framework format. Nucleic Acids Res 40:D453–D460 ArticleCASPubMedGoogle Scholar
- Koonin EV, Galperin MY (2003) Sequence - evolution - function: computational approaches in comparative genomics. Chapter 3, Information Sources for Genomics. Kluwer Academic, Boston. https://www.ncbi.nlm.nih.gov/books/NBK20256/
- Kulski JK (2016) Next-generation sequencing – an overview of the history, tools, and “Omic” applications, next generation sequencing-advances, applications and challenges. InTech
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35(6):1547–1549. https://doi.org/10.1093/molbev/msy096ArticleCASPubMedPubMed CentralGoogle Scholar
- Leinonen R, Akhtar R, Birney E, Bower L, Cerdeno-Tárraga A, Cheng Y, Cleland I, Faruque N, Goodgame N, Gibson R, Hoad G, Jang M, Pakseresht N, Plaister S, Radhakrishnan R, Reddy K, Sobhany S, Ten Hoopen P, Vaughan R, Zalunin V, Cochrane G (2011) The European nucleotide archive. Nucleic Acids Res 39:D28–D31 ArticleCASPubMedGoogle Scholar
- Lekamwasam S, Liyanage C (2013) Editorial. Galle Medical Journal 18(1). https://doi.org/10.4038/gmj.v18i1.5520
- Li MW, Qi X, Ni M, Lam HM (2013) Silicon era of carbon-based life: application of genomics and bioinformatics in crop stress research. Int J Mol Sci 14(6):11444–11483 ArticlePubMedPubMed CentralGoogle Scholar
- Lobo I (2008) Basic Local Alignment Search Tool (BLAST). Nature Education 1(1):215 Google Scholar
- Madeira F, Park YM, Lee J, Buso N, Gur T, Madhusoodanan N, Basutkar P, Tivey ARN, Potter SC, Finn RD, Lopez R (2019) The EMBL-EBI search and sequence analysis tools APIs. Nucleic Acids Res 47(W1):W636–W641. https://doi.org/10.1093/nar/gkz268ArticleCASPubMedPubMed CentralGoogle Scholar
- Magariños MP, Carmona SJ, Crowther GJ, Ralph SA, Roos DS, Shanmugam D, Van Voorhis WC, Agüero F (2012) TDR Targets: a chemogenomics resource for neglected diseases. Nucleic Acids Res (Database issue):D1118–D1127. https://doi.org/10.1093/nar/gkr1053
- Martins IM, Matos M, Costa R, Silva F, Pascoal A, Estevinho LM, Choupina AB (2014) Transglutaminases: recent achievements and new sources. Appl Microbiol Biotechnol 98:6957–6964 ArticleCASPubMedGoogle Scholar
- Martins IM, Meirinho S, Costa R, Cravador A, Choupina A (2019) Cloning, characterization, in vitro and in planta expression of a necrosis-inducing Phytophthora protein 1 gene npp1 from Phytophthora cinnamomi. Mol Biol Rep 46:6453–6462 ArticleCASPubMedGoogle Scholar
- Mehmood MA, Sehar U, Ahmad N (2014) Use of bioinformatics tools in different spheres of life sciences. J Data Mining Genomics Proteomics 5:158. https://doi.org/10.4172/2153-0602.1000158ArticleGoogle Scholar
- Mendez D, Gaulton A, Bento AP, Chambers J, De Veij M, Félix E, Magariños MP, Mosquera JF, Mutowo P, Nowotka M, Gordillo-Marañón M, Hunter F, Junco L, Mugumbate G, Rodriguez-Lopez M, Atkinson F, Bosc N, Radoux CJ, Segura-Cabrera A, Hersey A, Leach AR (2019) ChEMBL: towards direct deposition of bioassay data. Nucleic Acids Res 47(D1):D930–D940. https://doi.org/10.1093/nar/gky1075ArticleCASPubMedGoogle Scholar
- Mitra A, Kesarwani AK, Pal D, Nagaraja V (2011) WebGeSTer DB—a transcription terminator database. Nucleic Acids Res 39:129–135 ArticleGoogle Scholar
- Miyazaki S, Sugawara H, Gojobori T, Tateno Y (2003) DNA Data Bank of Japan (DDBJ) in XML. Nucleic Acids Res 31:13–16 ArticleCASPubMedPubMed CentralGoogle Scholar
- Münch R, Hiller K, Grote A, Scheer M, Klein J, Schobert M, Jahn D (2005) Virtual footprint and PRODORIC: an integrative framework for regulon prediction in prokaryotes. Bioinformatics 21:4187–4189 ArticlePubMedGoogle Scholar
- Nielsen H, Tsirigos KD, Brunak S, von Heijne G (2019) A brief history of protein sorting prediction. Protein J 38:200–216. https://doi.org/10.1007/s10930-019-09838-3ArticleCASPubMedPubMed CentralGoogle Scholar
- Okonechnikov K, Golosova O, Fursov M, the UGENE team (2012) Unipro UGENE: a unified bioinformaticstoolkit. Bioinformatics. 28(8):11667. https://doi.org/10.1093/bioinformatics/bts091ArticleCASGoogle Scholar
- Orchard S, Ammari M, Aranda B, Breuza L, Briganti L, Broackes-Carter F, Campbell NH, Chavali G, Chen C, del-Toro N, Duesbury M, Dumousseau M, Galeota E, Hinz U, Iannuccelli M, Jagannathan S, Jimenez R, Khadake J, Lagreid A, Licata L, Lovering RC, Meldal B, Melidoni AN, Milagros M, Peluso D, Perfetto L, Porras P, Raghunath A, Ricard-Blum S, Roechert B, Stutz A, Tognolli M, van Roey K, Cesareni G, Hermjakob H (2014) The MIntAct project-IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res 42(D1):D358–D363. https://doi.org/10.1093/nar/gkt1115ArticleCASPubMedGoogle Scholar
- Parra G, Blanco E, Guigó R (2000) GeneID in Drosophila. Genome Res 10(4):511–515. https://doi.org/10.1101/gr.10.4.511ArticleCASPubMedPubMed CentralGoogle Scholar
- Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M, Pérez E, Uszkoreit J, Pfeuffer J, Sachsenberg T, Yilmaz S, Tiwary S, Cox J, Audain E, Walzer M, Jarnuczak AF, Ternent T, Brazma A, Vizcaíno JA (2019) The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res 47(D1):D442–D450. https://doi.org/10.1093/nar/gky1106ArticleCASPubMedGoogle Scholar
- Potter SC, Luciani A, Eddy SR, Park Y, Lopez R, Finn RD (2018) HMMER web server: 2018 update. Nucleic Acids Res 46:W200–W204 ArticleCASPubMedPubMed CentralGoogle Scholar
- Raghava GPS (2002) APSSP2 : a combination method for protein secondary structure prediction based on neural network and example based learning. CASP5. A-132
- Rampp M, Soddemann T, Lederer H (2006) The MIGenAS integrated bioinformatics toolkit for web-based sequence analysis. Nucleic Acids Res 34:W15–W19. https://doi.org/10.1093/nar/gkl254ArticleCASPubMedPubMed CentralGoogle Scholar
- Resource Coordinators NCBI (2018) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 46(D1):D8–D13. https://doi.org/10.1093/nar/gkx1095ArticleCASGoogle Scholar
- Rodger S, David PJ, James KB (2003a) Analysing sequences using the Staden package and EMBOSS. In: Krawetz SA, Womble DD (eds) Introduction to Bioinformatics. A Theoretical and Practical Approach. Human Press Inc., Totawa, p 07512 Google Scholar
- Rodger S, David PJ, James KB (2003b) Managing sequencing projects in the GAP4 environment. In: Krawetz SA, Womble DD (eds) Introduction to Bioinformatics. A Theoretical and Practical Approach. Human Press Inc., Totawa, p 07512 Google Scholar
- Rost B, Sander C, Schneider R (1994) PHD--an automatic mail server for protein secondary structure prediction. Comput Appl Biosci 10:53–60 CASPubMedGoogle Scholar
- Salomon-Ferrer R, Case DA, Walker RC (2013) An overview of the Amber biomolecular simulation package. WIREs Comput Mol Sci 3:198–210 ArticleCASGoogle Scholar
- Sigrist CJA, de Castro E, Cerutti L, Cuche BA, Hulo N, Bridge A, Bougueleret L, Xenarios I (2012) New and continuing developments at PROSITE. Nucleic Acids Res 41:D344–D347. https://doi.org/10.1093/nar/gks1067ArticleCASPubMedPubMed CentralGoogle Scholar
- Spooner W, McLaren W, Slidel T, Finch DK, Butler R, Campbell J, Eghobamien L, Rider D, Kiefer CM, Robinson MJ, Hardman C, Cunningham F, Vaughan T, Flicek P, Huntington CC (2018) Haplosaurus computes protein haplotypes for use in precision drug design. Nat Commun 9:4128. https://doi.org/10.1038/s41467-018-06542-1ArticleCASPubMedPubMed CentralGoogle Scholar
- Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, Jensen LJ, Mering CV (2019) STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 47(D1):D607–D613. https://doi.org/10.1093/nar/gky1131ArticleCASPubMedGoogle Scholar
- UniProt Consortium (2008) The universal protein resource (UniProt). Nucleic Acids Res 36:D190–D195 ArticleGoogle Scholar
- Wang Y, Geer LY, Chappey C, Kans JA, Bryant SH (2000) Cn3D: sequence and structure views for Entrez. Trends Biochem Sci 25(6):300–302 ArticleCASPubMedGoogle Scholar
- Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, Lepore R, Schwede T (2018) SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46(W1):W296–W303 ArticleCASPubMedPubMed CentralGoogle Scholar
- Webb B, Sali A (2016) Comparative protein structure modeling using Modeller. Curr Protoc Bioinformatics 54:5.6.1–5.6.37 John Wiley, Sons, Inc. ArticleGoogle Scholar
- Weckx S, Del-Favero J, Rademakers R, Claes L, Cruts M, De Jonghe P, Van Broeckhoven C, De Rijk P (2005) novoSNP, a novel computational tool for sequence variation discovery. Genome Res 15:436–442 ArticleCASPubMedPubMed CentralGoogle Scholar
- Wishart DS, Feunang YD, Guo AC, Lo EJ, Marcu A, Grant JR, Sajed T, Johnson D, Li C, Sayeeda Z, Assempour N, Iynkkaran I, Liu Y, Maciejewski A, Gale N, Wilson A, Chin L, Cummings R, Le D, Pon A, Knox C, Wilson M (2018) DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. https://doi.org/10.1093/nar/gkx1037
- Yu CS, Cheng CW, Su WC, Chang KC, Huang SW, Hwang JK, Lu CH (2014) CELLO2GO: a web server for protein subCELlular LOcalization prediction with functional gene ontology annotation. PLoS One 9(6):e99368. https://doi.org/10.1371/journal.pone.0099368ArticleCASPubMedPubMed CentralGoogle Scholar
- Yunxia W, Song Z, Fengcheng L, Ying Z, Ying Z, Zhengwen W, Runyuan Z, Jiang Z, Yuxiang R, Ying T, Chu Q (2019) Therapeutic target database 2020: enriched resource for facilitating research and early development of targeted therapeutics. Nucleic Acids Res 48(D1):D1031–D1041. https://doi.org/10.1093/nar/gkz981 ISSN 1362-4962 ArticleCASGoogle Scholar
- Zhang S, Zhang L, Wang Y, Liao M, Bi S, Xie Z, Ho C, Wan X (2018) TBC2target: a resource of predicted target genes of tea bioactive compounds. Front Plant Sci 9:211. Published 2018 Feb 22. https://doi.org/10.3389/fpls.2018.00211ArticleCASPubMedPubMed CentralGoogle Scholar
Funding
The authors are grateful to the Foundation for Science and Technology (FCT, Portugal) and FEDER under Programme PT2020 for financial support to CIMO (UID/AGR/00690/2019).
Author information
Authors and Affiliations
- Centro de Investigação de Montanha (CIMO), Instituto Politécnico de Bragança, Campus de Santa Apolónia, 5300-253, Bragança, Portugal Iuliia Branco & Altino Choupina
- Iuliia Branco
You can also search for this author in PubMed Google Scholar
You can also search for this author in PubMed Google Scholar
Contributions
I.B. and A.C. designed prepared the manuscript; A.C. wrote the manuscript; and I.B. made the corrections including the English text and confirmation of the bibliography and URLs.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Human and animal rights
No human participants or animals were involved in this research.
Informed consent
This manuscript is original and submitted with the consent of all authors.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Branco, I., Choupina, A. Bioinformatics: new tools and applications in life science and personalized medicine. Appl Microbiol Biotechnol 105, 937–951 (2021). https://doi.org/10.1007/s00253-020-11056-2
- Received : 29 November 2020
- Revised : 29 November 2020
- Accepted : 09 December 2020
- Published : 06 January 2021
- Issue Date : February 2021
- DOI : https://doi.org/10.1007/s00253-020-11056-2
Share this article
Anyone you share the following link with will be able to read this content:
Get shareable link
Sorry, a shareable link is not currently available for this article.
Copy to clipboard
Provided by the Springer Nature SharedIt content-sharing initiative




